BUILDING UNIVERSAL STRATEGIES FOR STABILIZING PROTEIN COMPLEX STRUCTURE IN THE ABSENCE OF BULK SOLVENT.
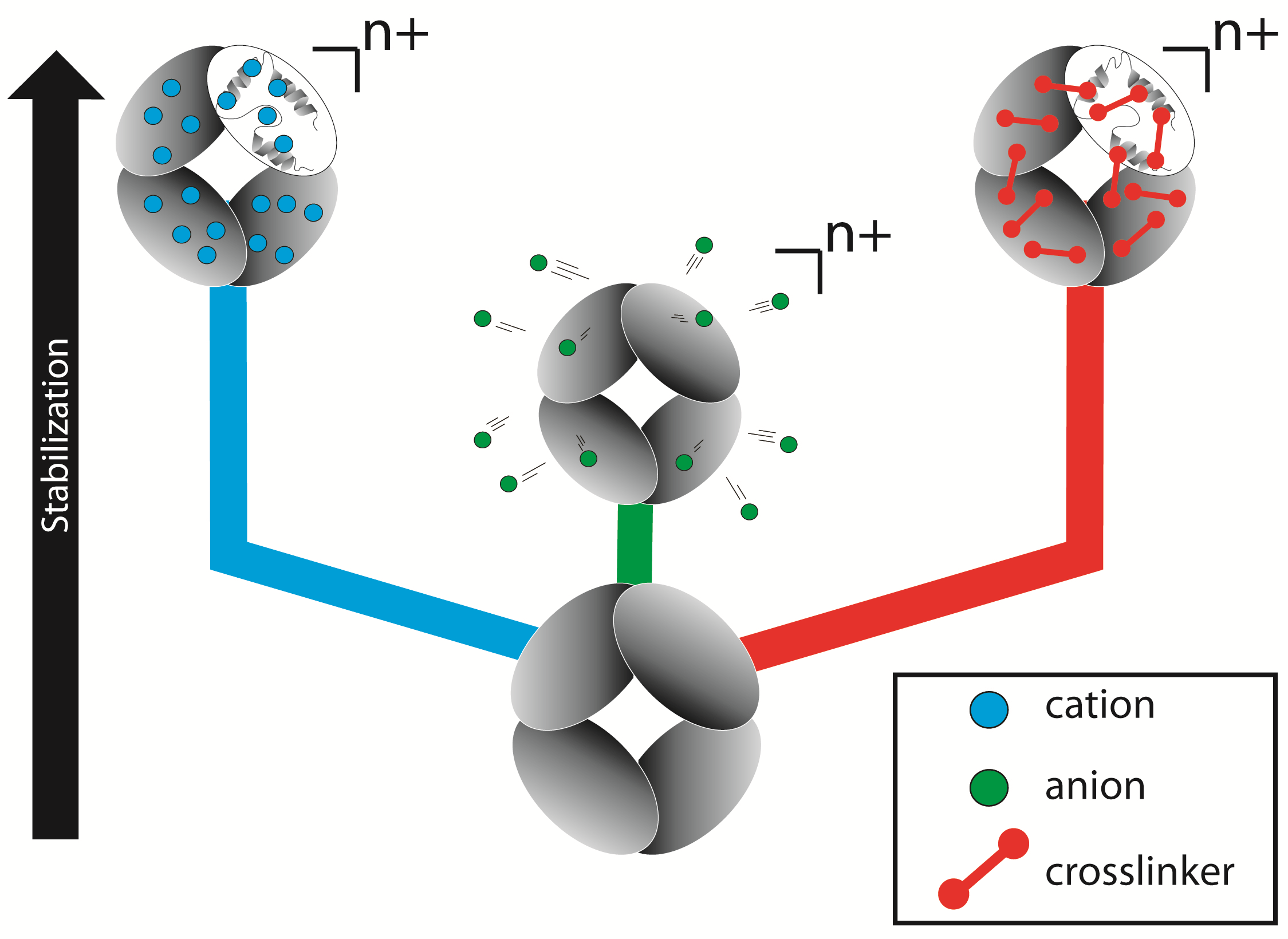
Ion mobility-mass spectrometry (IM-MS) is a powerful tool for structural biology with the ability to simultaneously assess the structure, topology, dynamics, and composition of large heterogeneous multiprotein complexes at low micromolar concentrations. However, one of the challenges in using IM-MS is that the structures of protein complexes in the gas phase can differ from those in solution as a consequence of ionization, desolvation, transport, and analysis over a range of time scale and energies. Thus, a general means must be identified to stabilize protein structures in the gas phase. Our approach involves stabilizing protein structure through the addition of inorganic cations and anions, or by crosslinking the intact complex. To quantitatively determine the increase in gas-phase protein stability, collision induced dissociation (CID) and collision induced unfolding (CIU) profiles of several multiprotein systems in the gas phase were generated on the Synapt G2 IM-MS platform (see reference 57 ). These results also suggest a mechanism for the added stabilization (see the Figure on this page) whereby anions, cations and crosslinkers stabilize gas-phase protein structures through different mechanisms. For example, anion-protein complexes exhibit primarily a "dissociative cooling" type mechanism characterized by the dissociation of protein-bound anions upon collisional activation (the green track, see reference 48), whereas cations tend to tightly bind to protein complexes and act to reduce Coulombic unfolding by tethering the flexible regions within protein ions through strong multi-dentate interactions (the blue track, see reference 56). Chemically crosslinking the protein complex creates covalent bonds across subunits, thus tethering protein regions together and preventing unfolding and dissociation (the red track, in collaboration with Phillip Andrews, UM Biological Chemistry).