| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
|
Human Fibroblast Growth Factor 12 (FHF 1) by Larry P. Taylor, Ph. D.
Feedback appreciated; please send comments to: Email: lpt Molecular & Behavioral Neuroscience Institute The University of Michigan Ann Arbor, MI |
My University Home Harris Links Chemistry / Modeling Links
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
Human Fibroblast Growth Factor 12 (FHF 1)
FGF-11, FGF-12, FGF-13, and FGF-14, also called FGF homologous factors FHF-3, FHF-1, FHF-2, and FHF-4, respectively, form a sub-group within the FGF family. Although this FGF sub-group does bind heparin (about the same magnitude as FGF 10, but significantly less than FGF 1 or FGF 2), the FHF group fails to bind to FGF receptors. There appears to be no competition between FGF and FHF proteins for binding to the FGF receptor. Instead, the FHF family binds directly to other proteins, specifically, an intra-cellular kinase scaffold protein, islet brain-2 (IB2).
Since the FHF family of protein ligands do not bind FGF receptors, the FHF sub-family of FGF ligands was recently removed from the FGF family. This change in "status" of the FHF sub-family of FGF ligands means that the FGF protein family is now composed of at least 18 homologous proteins that bind FGF receptors.
The unit cell for FGF 12 is
shown in Kinemage 1. The characteristics of the unit
cell for this structure are summarized at pdbsum.
The crystal structure shows four bound sulfate ions. (Kinemage 2
) Three of these are similar to other FGF heparin binding sites and most likely account for the fact that FGF 12 binds heparin. The probable residues involved in sulfate ion binding are Tyr-72, Gln-19, Lys-123, Arg-126, and Ser-133. A fourth sulfate resides in the vicinity of Arg-52, Lys-86 and Ser-88. Although not consistent with the strong heparin binding associated with the "basic canyon" of FGF 1 and FGF 2, this fourth site is similar to the sulfate site found in FGF 10.
Since the FHF family binds heparin, but does not bind to FGF receptors, it seems reasonable to assume there are surface features of the FHF family that interfere with binding to the FGF receptors. There are three likely locations for
steric interference which would limit the approach of FHF proteins to the FGF receptors.
(see Kinemage 3) The first area of divergence between FGF and FHF is the N-terminal region of FGF
12 (poorly resolved in the crystal). Based on an analogy to FGF 2 binding to FGFR 2, the FHF residues
6-9 (PQLK) would interfere with the
approach of the FHF protein to the D3 region of the FGF receptor.
The second possible conflict between FGF 12 and the FGF receptor centers around FGF 12 residue Tyr-93 in the area corresponding to the FGF low affinity loop (formed by beta sheets 8 and 9). If FGF 12 were to approach the FGF receptor in the same manner as FGF 1 and 2, then this Tyr-93 would extend into the D3 region of the receptor. This
contact clash limits binding possibilities. Other possible conflicts in this region are Arg-52
(Gly in FGF) and Val-95 (Asn in FGF). The Val in place of Asn is particularly
disrupting because the Asn residue in FGF is part of a network of hydrogen bonds between
the FGF ligand and the FGF receptor. The side chains of Valine do not form hydrogen bonds and thus, would be unable to
participate in a hydrogen bonding network.
The third possible area of conflict upon binding is the loop made beta sheets 9 and 10. The loop is 2 residues longer in FHF than
FGF. This would also interfere with approach of the protein to the D3 region of the FGF receptor.
The differences between FGF 12 and other FGF species is highlighted by superimposing the structure upon the universal FGF ligand FGF 2 (Kinemage 4).
The Kinemages:
The real-time visualization using KiNG of the structures on this site requires a java-enabled (JRE from Java) browser.
Possible Icons to the left of molecular model image on the download page
| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
A single click on the KiNG logo will launch the appropriate kinemage.
Kinemage 1: The FGF 12 Molecule
View 1 FGF 12
View 2 closer view of the FGF 12 low affinity loop region
|
224 K |
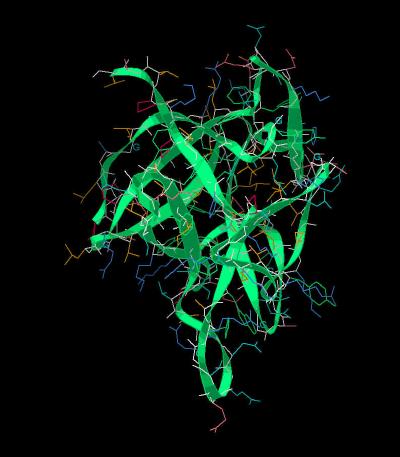 |
| Click on KiNG to see | FGF 12 |
Kinemage 2: The FGF 12 Sulfate Binding Residues
The probable residues involved in sulfate ion binding are Tyr-72, Gln-19, Lys-123, Arg-126, and Ser-133. A fourth sulfate resides in the vicinity of Arg-52, Lys-86 and Ser-88.
View 1 the 4 sulfate ions of the crystal
View 2 sites analogous to the "Basic Canyon"
View 3 sulfate binding region that is different from FGF 2
|
194 K |
 |
| Click on KiNG to see | Residues Binding Sulfates |
Kinemage 3: Differences Between FGF and FHF
Based on an analogy to FGF 2 binding to FGFR 2, the FHF residues
6-9 (PQLK) would interfere with the
approach of the FHF protein to the D3 region of the FGF receptor.
The second possible conflict between FGF 12 and the FGF receptor centers around FGF 12 residue Tyr-93 in the area corresponding to the FGF low affinity loop (formed by beta sheets 8 and 9). If FGF 12 were to approach the FGF receptor in the same manner as FGF 1 and 2, then this Tyr-93 would extend into the D3 region of the receptor. This
contact clash limits binding possibilities. Other possible conflicts in this region are Arg-52
(Gly in FGF) and Val-95 (Asn in FGF). The Val in place of Asn is particularly
disrupting because the Asn residue in FGF is part of a network of hydrogen bonds between
the FGF ligand and the FGF receptor. The side chains of Valine do not form hydrogen bonds and thus, would be unable to
participate in a hydrogen bonding network.
The third possible area of conflict upon binding is the loop made beta sheets 9 and 10. The loop is 2 residues longer in FHF than
FGF. This would also interfere with approach of the protein to the D3 region of the FGF receptor.
View 1 regions of difference between FGF and FHF
View 2 loop from beta sheets 9 and 10
View 3 low affinity loop region (Tyr-93)
|
192 K |
 |
| Click on KiNG to see | Residues Prohibiting Binding To FGF Receptors |
Kinemage 4: FGF 12 Superimposed on FGF 2
View 1 the overlay
View 2 loop area created by beta sheets 9 and 10 ... note the extended size of FGF 12 relative to FGF 2.
View 3 Tyr-93 (toggle side chains on to see the Tyr residue ... note
how it forms an extension not found in FGF 2)
|
291 K |
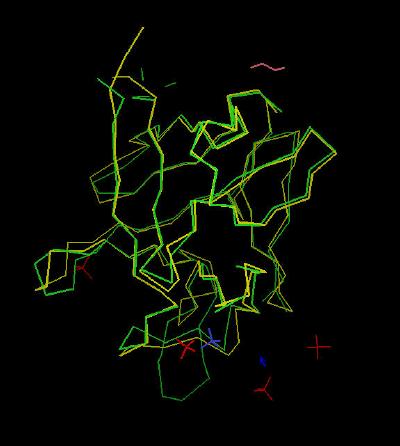 |
| Click on KiNG to see | FGF 12 Superimposed on FGF 2 |
Sequence: Residues 22-155 of the human FGF-12
Unresolved N-Terminal: RRRPE
X-Ray Resolved: PQLKGIVTRLFSQQGYFLQMHPDGTIDGTKDENSDYTLFNLIPVGLRVVAIQGVKASLYVAMNGEGYLYSSDVFTPEC
KFKESVFENYYVIYSSTLYRQQESGRAWFLGLNKEGQIMKGNRVKKTKPSSHFVPKPIEV
Unresolved C-Terminal: CMYREPSLHEIG
Source:
The DNA fragment encoding residues 1-181 was sub-cloned into pET-3 bacterial expression vector and expressed in E. coli. The structural coordinates are from Brookhaven Database file1Q1U. Cys-144 of the human sequence was mutated to Ala to prevent disulfide oligiomerization.
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
My University Home Harris Links Chemistry / Modeling Links
Copyright 2005-2020 by Larry P. Taylor
Molecular & Behavioral Neuroscience Institute
University of Michigan
All Rights Reserved
Supported by the Pritzker Neuropsychiatric Disorders Research Consortium, and by NIH Grant 5 P01 MH42251, Conte Center Grant #L99MH60398, RO1 DA13386 and the Office of Naval Research (ONR) N00014-02-1-0879 to Huda Akil & Stanley J. Watson. at the Molecular & Behavioral Neuroscience Institute.