| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
|
FGF/FGFR beta Sheet Nomenclature by Larry P. Taylor, Ph. D.
Feedback appreciated; please send comments to: Email: lpt Molecular & Behavioral Neuroscience Institute The University of Michigan Ann Arbor, MI |
My University Home Harris Links Chemistry / Modeling Links
FGF Site: FGF Intro Notes References FGF Sequences FGFR Sequences
beta Sheet Nomenclature
FGF molecules share a common protein architecture: the beta trefoil (three-fold repeat of a four-stranded sheet assembly without extended alpha helix strands possessing a pseudo three-fold axis of symmetry) motif. FGF ligands typically contain 12 beta sheets arranged in 3 groups of 4 strands. The C and N terminal sheets are adjacent which serves to close and stabilize the overall structure. Heparin binding to FGF stabilizes this junction and may play a key role in regulation of this molecule. The beta sheets are numbered 1 through 12, beginning at the N terminal of the sequence. The base of the trefoil (sheets 1, 4, 5, 8, 9, & 12) form a beta barrel motif. This architecture for FGF 2 is shown in
Kinemage 1.
Since Telokin has a 7 anti-parallel beta strand architecture common to
immunoglobulins, it serves as a reference template for the FGFR ligand binding
domains. The Telokin sheets are arranged into two primary parallel groups (of 4 and 3 strands) whose apparent wrapping around a central core has been described as a "beta barrel." The 7 sheets
of Telokin are described numerically in Kinemage 2
and labeled alphabetically from A-G in Kinemage 3.
The Telokin architecture is summarized at PDB
Sum.
The FGFR beta sheets have been mapped onto a Telokin template. This scheme, using the ligand binding domains D2 and D3 from the FGF 2-FGFR 1 dimer (Brookhaven 1CVS), is depicted numerically in Kinemage 4 and alphabetically in Kinemage 5.
Finally, the ligand binding domain is split into three defined immunoglobulin-like regions: domains D2, the
Linker (between D2 and D3) and domain D3. These are shown in Kinemage 6.
The Kinemages:
The real-time visualization using KiNG of the structures on this site requires a java-enabled (JRE from Java) browser.
Possible Icons to the left of molecular model image on the download page
| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
A single click on the KiNG logo will launch the appropriate kinemage.
Kinemage 1: The FGF beta Sheets
View 1 the sheets identified
View 2 the molecule rotated to highlight the three distinctive folds of the trefoil architecture. This arrangement is maintained by the fold (in
red) in which the two beta sheets 1 (N-terminal) and 12 (C-terminal) bring together the loose terminal ends of the protein sequence. The base of the FGF trefoil (sheets 1,4,5,8,9,& 12) form a beta barrel motif.
|
298 K |
 |
| Click on KiNG to see | FGF Sheets |
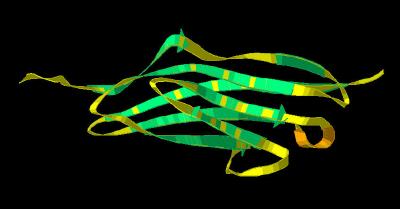
Kinemage 2: Telokin Template (Numerical)
View 1 identifies the 7 anti-parallel beta sheets as numbered
sheets
View 2 an arbitrary side view
View 3 an arbitrary top view
View 4 the "vertical" view
|
188 K |
 |
| Click on KiNG to see |
Telokin Sheets (Numerical) |
Kinemage 3: Telokin Template (Alphabetical)
Telokin is a classic representation of the immunoglobin-like "beta barrel" architecture.
View 1 identifies the 7 anti-parallel beta sheets
as English alphabetical characters
View 2 an arbitrary side view
View 3 an arbitrary top view
View 4 the "vertical" view
|
187 K |
 |
| Click on KiNG to see | Telokin Sheets (Alphabetical) |
Kinemage 4: FGFR Ligand Binding Domain beta
Sheets (Numerical)
The secondary structure of domains D2-D3 of the FGFR ligand binding domain are mapped onto a Telokin template using numerical designations.
View 1 FGFR 1 ligand binding domain
View 2 the "vertical" view
|
395 K |
 |
| Click on KiNG to see | FGFR Sheets (Numerical) |
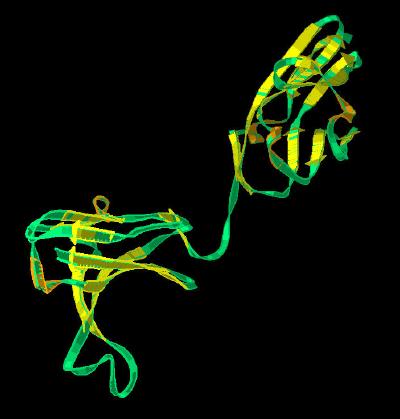
Kinemage 5: FGFR Ligand Binding Domains (Alphabetical)
The secondary structure of domains D2-D3 of the FGFR ligand binding domain are mapped onto a Telokin template using alphabetical designations.
View 1 FGFR 1 ligand binding domain
View 2 the "vertical" view
|
394 K |
 |
| Click on KiNG to see | FGFR Sheets (Alphabetical) |
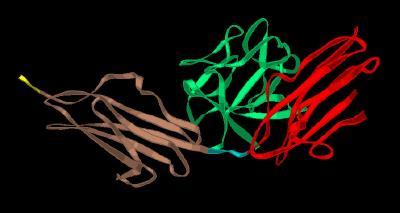
Kinemage 6: FGFR Ligand Binding Domains
This is a simple schematic to highlight the D2, linker and D3 Domains of FGFR's.
View 1 FGF-FGFR Complex
View 2 "Vertical" view often seen in journals
|
416 K |
 |
| Click on KiNG to see | FGFR Domains |
Sources:
FGF 2 Coordinates were taken from Brookhaven Database file 1BFF. Telokin coordinates were taken from Brookhaven Database file 1FHG. FGFR coordinates were taken from Brookhaven Database file 1CVS.
FGF Site: FGF Intro Notes References FGF Sequences FGFR Sequences
My University Home Harris Links Chemistry / Modeling Links
Copyright 2005-2020 by Larry P. Taylor
Molecular & Behavioral Neuroscience Institute
University of Michigan
All Rights Reserved
Supported by the Pritzker Neuropsychiatric Disorders Research Consortium, and by NIH Grant 5 P01 MH42251, Conte Center Grant #L99MH60398, RO1 DA13386 and the Office of Naval Research (ONR) N00014-02-1-0879 to Huda Akil & Stanley J. Watson. at the Molecular & Behavioral Neuroscience Institute.