| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
|
by Larry P. Taylor, Ph. D.
Feedback appreciated; please send comments to: Email: lpt Molecular & Behavioral Neuroscience Institute The University of Michigan Ann Arbor, MI |
My University Home Harris Links Chemistry / Modeling Links
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
Human FGF 9 Monomer
FGF 9 was originally described as glia-activating factor (GAF). It shares about 30% sequence identity with FGF 2. FGF 9 activates proliferation of glia cells without being mitogenic for endothial cells. The specificity of biological activity is most likely related to its differential receptor binding profile. Its highest binding affinity is with the FGF 3 (IIIc) receptor.
Like other FGF type molecules FGF 9 show the beta trefoil structure. (Kinemage 1
, Kinemage 2 , Kinemage 3 ). The backbone of FGF 9 is superimposed on the backbone of the prototypical FGF molecule, FGF 2, in
Kinemage 4. There are a few distinct differences between FGF 2 and FGF 9. At FGF 9 position Thr-70 to Gly-71
(Kinemage 4, view 2 ... an extra glycine here in FGF 2 adds a one residue length to the FGF 2 loop; bottom, left of center) the FGF 9 loop is one residue shorter than the corresponding area in FGF 2. The FGF 9 loop at Asp-88 to Ser-90 shows an altered orientation
(Kinemage 4, view 2: right) compared to FGF 2. A major divergence between FGF 2 and FGF 9 occurs in the vicinity of Tyr-153 to Arg-161 (the low affinity loop,
Kinemage 4, view 3; bottom left). Since this is a primary contact point between FGF ligands and receptors, this divergence in structure is most likely a prime component in FGF 9 receptor selectivity. Lastly, the N and C-terminal regions
(Kinemage 4, view 1 & 5) of FGF 9 show a substantial extension beyond the beta trefoil core, relative to FGF 2, of alpha helix secondary structure. This is a major departure from most FGF molecules which have X-Ray unresolved C and N-terminal regions. So. the well-ordered secondary structure of the FGF 9 molecule be considered as a separate entity from the FGF beta trefoil core.
The characteristics of the unit cell for this structure are summarized at pdbsum.
The crystal structure for FGF 2 and FGF 9 also show different locations for the trapped phosphate ion (assumed to be potential interaction areas for heparin sulfates). The FGF 2 crystal structure shows one phosphate ion in the same volume commonly associated with the sulfate ion and heparin binding. The FGF 9 molecule
(Kinemage 4, view 1) also has a phosphate bound in this region, but traps an additional phosphate ion in a region not traditionally associated with heparin binding. This is most likely a result of interaction with residues Arg-160 and Arg-180.
(Kinemage 5 )
FGF 9, perhaps more than any FGF moiety, shows a tendency to self-dimerize under physiological conditions. Since other FGF molecules do not have the well-ordered alpha helix C and N terminal extensions, these extensions appear to be a significant contributor to the tendency to form dimers in the absence of heparin. (Other FGF species typically show structures that are X-ray unresolved at the extensions. The inability to resolve the extensions is associated with a lack of organized structure,)
The primary driving force for dimerization is the exclusion of solvent via multiple hydrophobic contacts of the alpha helical terminal extension region. These involve residues Leu-54, Leu-57, Ile-60, Leu-61, Pro-194, Val-197 and Leu-200 of both chains involved in the dimerization. These are shown in
Kinemage 6.
Although the FGF 9 dimer interface is hall marked by hydrophobic interactions, there is some stability to dimerization added by 4 hydrogen bonds and 2 salt bridges. There are hydrogen bonds between each of the C and N terminal helix extensions at Arg-64 to the backbone carbonyl of Val-197 as well as at beta trefoil residues Tyr-67 to Asn-143. There are salt bridges between each Arg-62 and Asp-193 combination of the dimer pair. The individual residues involved are shown in
Kinemage 6. These interactions in the dimer are shown in the kinemage of the FGF 9 dimer structure (1G82).
The FGF ligands contact three regions of the FGF receptor ( Kinemage
7 ). It is assumed that conserved residues in the contact regions are critical for strong binding interactions. The FGF 9 residues that would be important for maintaining the primarily hydrophobic contacts with the D2 region of the of a FGF receptor are Tyr-47, Tyr-145, and Leu-188.
Consistent with other FGF ligands, the FGF 9 residue, Asn-146, forms a hydrogen bond to an invariant Arginine residue of the receptor linker region (Arg-250 in FGFR 1).
The FGF 9 region contacting the D3 region of the FGF receptor bears little sequence resemblance to other FGF ligands. So, this can be considered the primary region of selectivity of FGF 9 for the FGF 3 (IIIc) receptor. The FGF 9 contact residues for the FGF receptor D3 domain are Ile-98, Ile-100, Tyr-115, Leu-130 and Val-135. This arrangement of hydrophobic residues clashes with the hydophobics of the D3 domain of most FGF receptors, but fits into the geometry of the FGFR 3 (IIIc).
There are two phosphate ions trapped in the crystal structure (Kinemage
5 ). Since biological heparin is typically longer than the dodecyl sugar polymer commonly used to used the FGF-FGFR complex, it is reasonable to assume both areas binding phosphate could be involved in binding a heparin helix in the FGF-heparin-FGFR complex. In addition, a "distal" site (corresponding to a sulfate ion site in other FGF species) centered around Thr-81, Lys-121 and Glu-123 most likely is also involved in heparin
binding. The primary site (corresponding to the "basic canyon" in other FGF species) is highlighted by residues Arg-69, Tyr-153, Tyr-163, Lys-168, Arg-173, Arg-177 and Arg-180. The multiple sites
(Kinemage 8 ) probably allow some a form of regulation
of heparin (or
FGF-heparin-FGFR complex) binding.
An interesting observation is that in the FGF 9 dimer, many of the potential FGF to FGF receptor interaction sites are unavailable to the receptor because they are buried within the FGF 9 dimer. This is seen by examining the FGF 9 dimer crystal structure
( FGF 9 (Dimer)).
The Kinemages
The real-time visualization using KiNG of the structures on this site requires a java-enabled (JRE from Java) browser.
Possible Icons to the left of molecular model image on the download page
| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
A single click on the KiNG logo will launch the appropriate kinemage.
Kinemage 1: The Calpha trace of the Unit Cell
|
14 K |
 |
| Click on KiNG to see | FGF 9 Crystal Structure |
Kinemage 2: Ribbon Rendering of FGF 9
|
262 K |
 |
| Click on KiNG to see | FGF 9 Monomer |
Kinemage 3: Secondary Structure Cartoon of FGF 9
|
189 K |
 |
| Click on KiNG to see | FGF 9 Cartoon |
Kinemage 4: Superimposition of FGF 9 upon FGF 2
FGF 9 was superimposed upon FGF 2 (1BFF) with Deep View
"Magic Fit." Non-protein material from the 1BFF was included to
maintain orientation during image manipulation.
The individual FGF chains were extracted from the unit cell (IG82) with SYBYL and then superimposed upon FGF 2 (1BFF) with Deep View "Magic Fit." Non-protein material from the 1BFF was included to maintain orientation during image manipulation.
View 1 the overlay
View 2 smaller loop at FGF 9 Arg-69 thru Gly-71
View 3 alteration at FGF 9 Asp-88 thru Ser-90 loop
View 4 low affinity loop
View 5 Glu-142
View 6 N & C terminal junction
|
318 K |
 |
| Click on KiNG to see | FGF 9 Upon FGF 2 |
Kinemage 5: The FGF 9 Additional phosphate site
The "distal" site (corresponding to a sulfate ion site in other FGF species) centered around Thr-81, Lys-121 and Glu-123 most likely is also involved in heparin binding. The primary site (corresponding to the "basic canyon" in other FGF species) is highlighted by residues Arg-69, Tyr-153, Tyr-163, Lys-168, Arg-173, Arg-177 and Arg-180.
View 1 FGF 9
View 2 the extra phosphate ion highlighting potential interacting residues Arg-160 and Arg-180.
|
224 K |
 |
| Click on KiNG to see | The FGF 9 Additional Phosphate Site |
Kinemage 6: FGF 9 Dimer Contact Points
There are hydrogen bonds between each of the C and N terminal helix extensions at Arg-64 to the backbone carbonyl of Val-197 as well as at beta trefoil residues Tyr-67 to Asn-143. There are salt bridges between each Arg-62 and Asp-193 combination of the dimer pair.
View 1 FGF 9
View 2 primary hydrophobic points Leu-54, Leu-57, Ile-60, Leu-61, Pro-194, Val-197 and Leu-200.
View 3 hydrogen bonding pairs Arg-64 to backbone carbonyl of Val-197 as well as at beta trefoil residues Tyr-67 to Asn-143.
View 4 salt bridge component residues of Arg-62 and Asp-193.
|
226 K |
 |
| Click on KiNG to see | The Dimer Contact Residues |
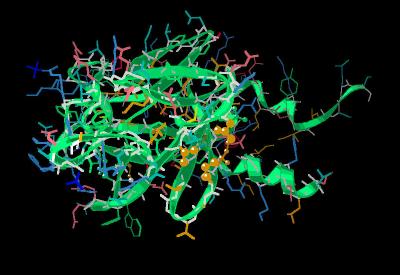
Kinemage 7: FGF 9- Receptor D2 Contact Points
The FGF 9 residues maintaining the primarily hydrophobic contacts with the D2 region of the of a FGF receptor are Tyr-47, Tyr-145, and Leu-188.
View 1 the ligand
View 2 D2 contact points Tyr-47, Tyr-145, and Leu-188.
View 3 D2-D3 linker contact Asn-156.
View 4 D3 contact points Ile-98, Ile-100, Tyr-115, Leu-130 and Val-135.
|
225 K |
 |
| Click on KiNG to see | FGF 9-Receptor D2 Contact Points |
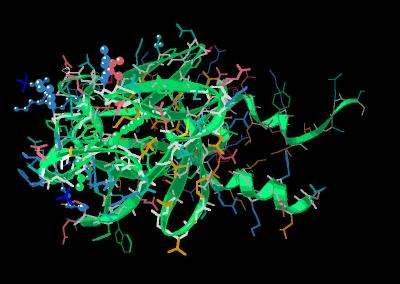
Kinemage 8: Heparin Contact Points
The "distal" site (corresponding to a sulfate ion site in other FGF species) centered around Thr-81, Lys-121 and Glu-123 most likely is involved in heparin binding. The primary site (corresponding to the "basic canyon" in other FGF species) is highlighted by residues Arg-69, Tyr-153, Tyr-163, Lys-168, Arg-173, Arg-177 and Arg-180.
View 1 FGF 9 ligand
View 2 distal heparin binding region centered around Thr-81, Lys-121 and Glu-123.
View 3 primary "basic canyon" residues of Arg-69, Tyr-153, Tyr-163, Lys-168, Arg-173, Arg-177 and Arg-180.
|
226 K |
 |
| Click on KiNG to see | Heparin Contact Points |
The Sequence
Unresolved N-Terminal: DHLGQSEAGGLPRGPAV
X-ray resolved: TDLDHLKGILRRRQLYCRTGFHLEIFPNGTIQGTRKDHSRFGI
LEFISIAVGLVSIRGVDSGLYLGMNEKGELYGSEKLTQECVFREQFEENWYNTYSSNLYK
HVDTGRRYYVALNKDGTPREGTRTKRHQKFTHFLPRPVDPDKVPELYKDILSQS
Source:
The first 33 residues of the gene sequence were not used because these residues are implicated in secretion, not ligand activity.
cDNA corresponding to human FGF 9 (residues 35-208) was expressed in E. coli. The structural coordinates were taken from the Brookhaven Database file
1IJK.
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
My University Home Harris Links Chemistry / Modeling Links
Copyright 2005-2020 by Larry P. Taylor
Molecular & Behavioral Neuroscience Institute
University of Michigan
All Rights Reserved
Supported by the Pritzker Neuropsychiatric Disorders Research Consortium, and by NIH Grant 5 P01 MH42251, Conte Center Grant #L99MH60398, RO1 DA13386 and the Office of Naval Research (ONR) N00014-02-1-0879 to Huda Akil & Stanley J. Watson. at the Molecular & Behavioral Neuroscience Institute.