| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
|
The FGF 1 - FGFR 2c - Heparin (2:2:1) Complex by Larry P. Taylor, Ph. D.
Feedback appreciated; please send comments to: Email: lpt Molecular & Behavioral Neuroscience Institute The University of Michigan Ann Arbor, MI |
My University Home Harris Links Chemistry / Modeling Links
FGF Site: FGF Intro Nomenclature Notes References FGF Sequences FGFR Sequences
The FGF 1 - FGFR 2 - Heparin (2:2:1) Complex
The crystal structure 1E0O shows a 2:2:1: arrangement of FGF 1, FGFR 2 and heparin. This can be viewed either as two 1:1 ligand-receptor complexes assembled via a common heparin molecule or as a single pentamer assembly. In the ligand-receptor complex (chains A and B), both components of the assembly contact the heparin decasaccharide, while in the other (chains C and D), only the FGF ligand (Chain C) contacts the heparin. Each component of the ligand-receptor complex shows a different mode of contact between the FGF 1 ligand and the sugar poly anion
heparin. The characteristics of the unit cell for this structure are summarized at pdbsum.
The two receptor chains (B and D) show different angles across the linker region between the their respective D2 and D3 domains. This is illustrated in
Kinemage 8. In this kinemage, the receptor chains were each separately superimposed (using
Deep View's Magic Fit) upon FGF 1 from a different crystal structure (1AXM).
The FGF 1 ligand contacts the receptor ligand-binding region comprised of an immunoglobulin-like domains D2 (I class; residues 152-249), a short linker chain (residues 250-253) and the
D3 domain (C2
class; residues 254-360). These receptor domains definitions are shown in Kinemage
6.
In this crystal structure, the heparin helix shows a distortion at sugar residues 2-4
( Kinemage 1 & Kinemage 8).
This distortion has not been observed in other FGF-FGFR complexes. The heparin-receptor contact points are residues Asn-18, Lys-112. Lys-113, Asn-114, Lys-118, Arg-119, Arg-122, Gln-127, Lys-128. These are shown in
Kinemage 7 . The two FGF 1 ligands in this crystal structure complex have different heparin contact points. The difference in spatial orientation of FGF 1 ligand to heparin is shown
Kinemage 9 & Kinemage 10. The ligand (chain C) is rotated about 120 degrees relative to the chain A ligand (about a pseudo C3 rotational axis) with respect to its heparin interaction. This allows a conserved residue, Trp-107, to become involved in the heparin binding. In addition, residues close to Trp-107 (Lys-105 and Pro-121) probably contribute to the stability of the FGF 1-heparin interaction. A side-by-side comparison of the two FGF-heparin binding combinations is show in
Kinemage 9, while the crystal position of the FGF 1 dimer (from 1E0O) relative to the ligand dimer pair of 1AXM is shown in
Kinemage 10. The heparin-receptor basic residue electrostatic contact points (Lys-164, His-167, Lys-176, and Arg-178) and Van der Waal's contact points (Thr-104 and Val-175) are shown in
Kinemage 11.
The primary cause for difference in the D3 orientation with respect to the D2 domain appears to be residue Pro-253. This residue can undergo cis-trans isomerism and, as such, can alter the relative positions of the
receptor domains with respect to each other.
This hinge region surrounding this residue can be viewed in the receptor comparisons of
Kinemage 8. The Pro-253 residue is highlighted in Kinemage
11,
The hinge region may represent a control point for biological activity.
The FGFR 2c heparin binding region centers around the FGF receptor conserved sequence MMEKRLHAVPAANVKFR (residues 161-178). All of the basic residues (except for Arg-165, which is oriented away from the heparin molecule) in this sequence interact with negatively charged sulfate or carboxylate groups of the heparin molecule. The 6-O-sulfate of GlcN is involved in several different interactions including hydrogen bonding to Lys-164,
His-167, and the backbone amide of Lys-176. There are also Van der Waal's contacts between this sulfate and Thr-174 and Val-175 of the receptor,
The FGF 1 ligand to the FGFR 2c D2 domain interactions are primarily hydrophobic. They primarily involve ligand residues Tyr-15, Gly-20, Phe-22, Tyr-94.
Leu-133 and Leu-135. Leu-133 and Leu-135 also form part of the hydrophobic pocket surrounding receptor linker region residue Arg-251. The D2 receptor residues involved in ligand binding are Lys-164, Leu-166, Ala-168, Val-169, and Pro-170. There are electrostatic interactions (enhanced by the surrounding hydrophobicity) between Arg-35 of the ligand to Glu-163 of the receptor. A similar salt bridge occurs between Arg-37 of the ligand and Asp-247 of the receptor.
The FGFR 2c linker region contains the critical residue, Arg-251. This receptor amino acid is surrounded by a hydrophobic pocket defined by ligand residues Leu-89, Leu-133 and Pro-134. This encircling volume of hydrophobicity enhances potential electrostatic and hydrogen bonding interactions. There is hydrogen bonding between the guanidinium group of receptor Arg-251 to the backbone carbonyl of ligand residue His-93. The combination of hydrophobic environment and the hydrogen bonding to His-93 places receptor Arg-251 in a position to form a strong hydrogen bond to ligand residue Asn-95. It has been shown that replacement of this FGF 1 residue Asn-95 with an alanine residue results in a 400 fold reduction in binding affinity.
The ligand to Domain D3 interactions center around a salt bridge between receptor conserved residue Arg-255 and ligand residue Glu-87. The Glu-87 position is stabilized by a hydrogen bond between Glu-87 and Tyr-97. The receptor conserved residue Ile-257 hydrophobically packs against ligand residues Leu-89 and His-93. This residue also lies along the backbone between ligand residues Glu-90 and Glu-91.
The only FGFR 2c splice-form variant interaction with FGF 1 occurs at ligand residue Val-51. This hydrophobic contact point interacts with receptor residues Ile-350 and Phe-352. The presence of only a single splice-form contact in the
splice variant region of D3 suggests a lack of splice-form specificity in FGF 1 to
FGFR 2c binding.
The ligand-receptor interactions are depicted in Kinemages 12.
The Kinemages
The real-time visualization using KiNG of the structures on this site requires a java-enabled (JRE from Java) browser.
Possible Icons to the left of molecular model image on the download page
| Java Not Activated | Java Not Activated | Java Functional |
 |
Blank Area
or message: Image requires a Java enabled browser
|
 |
| KiNG Inactive | KiNG Inactive | KiNG Full Functional |
A single click on the KiNG logo will launch the appropriate kinemage.
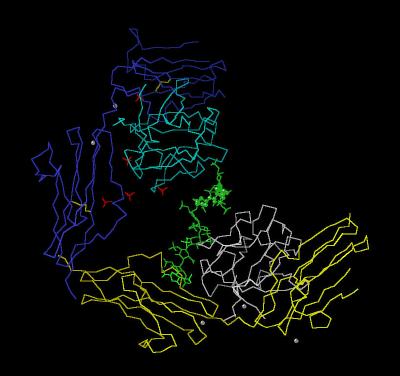
Kinemage 1: The FGF 1-FGFR 2c-Heparin (2:2:1) Complex
Classic Calpha trace of the FGF 1-FGFR 2c Complex with one heparin. The point of heparin distortion is marked with green spheres.
View 1 the 2:2:1 complex
View 2 closer look at the heparin distortion
|
53 K
|
 |
| Click on KiNG to see | The 2:2:1 Complex |
Kinemage 2: The FGF 1-FGFR 2c-Heparin (2:2:1) Complex
Ribbon Rendering
|
1,090 K |
 |
| Click on KiNG to see | Ribbon Rendering for the 2:2:1 Complex |
Kinemage 3: The FGF 1-FGFR 2c-Heparin (2:2:1) Complex: Ribbon Rendering 2
Here, only one FGF-FGFR unit has side chains colored by property.
|
1,086 K |
 |
| Click on KiNG to see | Only 1 FGF-FGFR Pair With All Additional Properties |
Kinemage 4: The FGF 1-FGFR 2-Heparin (2:2:1) Complex Secondary Structure Cartoon
|
844 K |
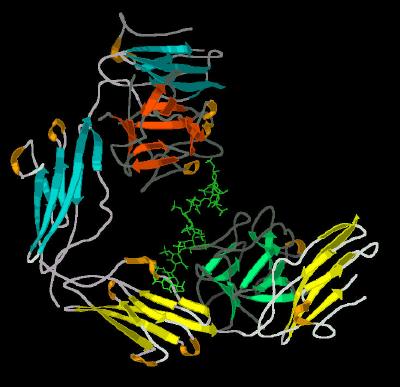 |
| Click on KiNG to see | Secondary Structure Cartoon for 2:2:1 Complex |
Kinemage 5: The FGF 1-FGFR 2c-Heparin (2:2:1) Complex Secondary Structure Cartoon
Kinemage 3 with secondary structure ribbon
|
841 K |
 |
| Click on KiNG to see | Secondary Structure Cartoon for 2:2:1 Complex |
Kinemage 6: Receptor Ligand Binding Domains
The three defined immunoglobulin-like ligand binding domains D2 (residues 152-249), the Linker (residues 259-253) and D3 ( residues 254-360)
|
1059 K |
 |
| Click on KiNG to see | The Domains Defined |
Kinemage 7:The FGF 1 to Heparin Contact Points
The FGF 1 to heparin contact points are residues Asn-18, Lys-112. Lys-113, Asn-114, Lys-118, Arg-119, Arg-122, Gln-127, Lys-128.
View 1 the complex
View 2 closer view looking down the heparin helix
View 3 closer view looking towards the sides of the heparin helix
|
1,075 K |
 |
| Click on KiNG to see | Heparin Contact Points |
Kinemage 8: Comparison of the FGF 1 & FGFR 2c Structures in Crystal Structures 1AXM and 1E0O
View 1 End-On
View 2 Top
View 3 Side
View 4 Bottom
View 5 the Proline Hinge (Pro 253). To visualize the hinge region::
Toggle Calphas Off
Toggle Main Chain On
Toggle Side Chains On
|
261 K |
 |
| Click on KiNG to see | 1AXM and 1E0O Comparisons |
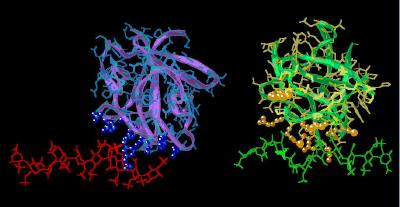
Kinemage 9: Comparison of the FGF 1 Structure in Crystal Structures 1AXM and 1E0O
Residues associated with heparin binding (Asn-18, Trp-107, Lys-112. Lys-113, Asn-114, Lys-118, Arg-119, Arg-122, Gln-127, Lys-128) are shown to aid in recognizing the different orientation of the two ligands with respect to heparin. Note how the rotation of Chain C (relative to Chain A) allows Trp-107 of chain C to become involved with the heparin.
View 1 the comparison
View 2 close up of Chain A-heparin contact region
View 3 close up of chain C-heparin contact region
|
313 K |
 |
| Click on KiNG to see | Comparison of the Heparin Sites |
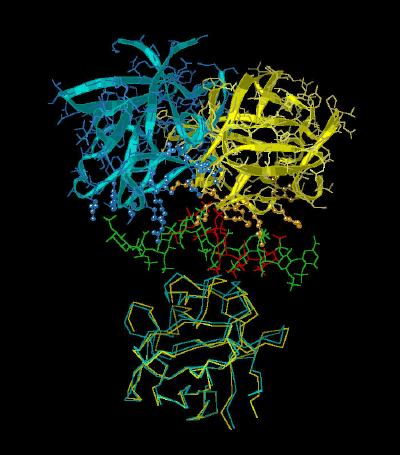
Kinemage 10: Positions of the FGF-Heparin-FGF Complex between the Crystal Structures of 1E0O and 1AXM
View 1 the tilt of the 1E0O complex relative to the similar arrangement in crystal structure 1AXM
View 2 close up of the heparin binding domain of 2AXM
View 3 close up of the heparin binding domain of 1E0O.
|
317 K |
 |
| Click on KiNG to see | Comparison of the FGF Positions |
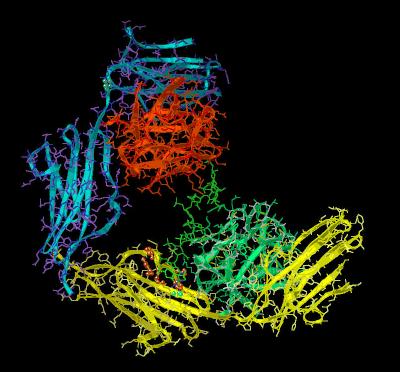
Kinemage 11: The Receptor-Heparin Binding Region
The heparin binding region of the receptor. is defined by residues 161-178, MMEKRLHAVPAANVKFR.
View 1 the 2:2:1 complex
View 2 closer view (arbitrary "end") of the heparin binding region of FGFR 2c
Chain B
View 3 closer view (arbitrary "side") of the heparin binding region of FGFR
2c Chain B
View 4 centers on the multiple binding interactions between heparin sulfate and FGFR 2c
Chain B.
|
1,046 K |
 |
| Click on KiNG to see | Receptor Heparin Contact Points |
Kinemage 12: Protein Ligand-Receptor Interactions
The hydrophobic ligand residues are Tyr-15, Gly-20, Phe-22, Tyr-94. Leu-133 and Leu-135. Leu-133 and Leu-135 and receptor residues Lys-164, Leu-166, Ala-168, Val-169, and Pro-170. There are electrostatic interactions between Arg-35 of the ligand to Glu-163 of the receptor. A similar salt bridge occurs between Arg-37 of the ligand and Asp-247 of the receptor.
The ligand-receptor linker region interaction centers around receptor residue Arg-251. This receptor amino acid is surrounded by a hydrophobic pocket defined by ligand residues Leu-89, Leu-133 and Pro-134.
The Ligand to Domain D3 interactions centers around receptor Arg-255 and ligand residue Glu-87. The Glu-87 position is stabilized by a hydrogen bond with Tyr-97. The receptor conserved residue Ile-257 hydrophobically packs against ligand residues Leu-89 and His-93. This residue also lies along the backbone between ligand residues Glu-90 and Glu-91.
A splice-form variant interaction with FGF 1 occurs at Val-51. This hydrophobic contact point interacts with receptor residues Ile-350 and Phe-352.
View 1 the 2:2:1 Complex
View 2 close up of the FGFR 2c Receptor Domain D2 Interactions with the FGF 1 Ligand
View 3 close up of the FGFR 2c Receptor Domain Linker Interactions with the FGF 1 Ligand.
View 4 close up of the FGFR 2c Receptor Domain D3 Interactions with the FGF 1 Ligand
View 5 close up of the isoform specific contact point between ligand and receptor.
|
932 K |
|
| Click on KiNG to see | Ligand-Receptor Contact Points |
Sequences:
Chain A:
FNLPPGNYKKPKLLYCSNGGHFLRILPDGTVDGTRDRSDQHIQLQLSAES
VGEVYIKSTETGQYLAMDTDGLLYGSQTPNEECLFLERLEENHYNTYISK
KHAEKNWFVGLKKNGSCKRGPRTHYGQKAILFLPLPVSSD
Chain B:
SNNKRAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKE
FKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLD
VVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKN
GSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYTCLAGNS
IGISFHSAWLTVLPAPGRE
Chain C:
FNLPPGNYKKPKLLYCSNGGHFLRILPDGTVDGTRDRSDQHIQLQLSAES
VGEVYIKSTETGQYLAMDTDGLLYGSQTPNEECLFLERLEENHYNTYISK
KHAEKNWFVGLKKNGSCKRGPRTHYGQKAILFLPLPVSSD
Chain D:
SNNKRAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKE
FKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLD
VVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKN
GSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYTCLAGNS
IGISFHSAWLTVLPAPGRE
Source:
Reconstituted human FGFR 2c (i.e. splice variant IIIc) with human FGF 1 and heparin decasaccharide in 2:2:1 mix; Structural coordinates from the Brookhaven Database File1E0O.
Top
FGF Site: FGF Intro
Nomenclature Notes
References FGF
Sequences FGFR Sequences
My University Home Harris Links Chemistry / Modeling Links
Copyright 2005-2020 by Larry P. Taylor
Molecular & Behavioral Neuroscience Institute
The University of Michigan
All Rights Reserved
Supported by the Pritzker Neuropsychiatric Disorders Research Consortium, and by NIH Grant 5 P01 MH42251, Conte Center Grant #L99MH60398, RO1 DA13386 and the Office of Naval Research (ONR) N00014-02-1-0879 to Huda Akil & Stanley J. Watson. at the Molecular & Behavioral Neuroscience Institute.