Our research covers genetics, systematics, and ecology of fungi. Our primary ongoing projects are:
- Role of mitotic recombination in pathogen adaptation
- Evolution of mating systems and nuclear behavior in mushroom fungi (Agaricomycetes)
- Population genetics of the fungal disease of amphibians (chytridiomycosis)
- Molecular systematics of early diverging fungal lineages (Chytridiomycetes and Zygomycetes sensu lato)
Evolution of mitotic recombination
Diploid pathogens ubiquitously undergo mitotic recombination (MR). Most perspectives on MR suggest negative effects on the phenotype, as MR is associated only with loss of heterozygosity rather than gain of genetic diversity. Yet, MR can alter multilocus genotypes, altering epistatic interactions among loci, and can increase the spread of beneficial alleles that are recessive. Why pathogens are so frequently observed to undergo LOH is the subject of our project in which we are using an experimental evolution approach through in vitro evolution and a yeast-insect pathogenesis model. Our results show that LOH can be adaptive and provide a rapidly accessed source of genotypic variation (James et al. 2019).
Genetics of mushroom sex
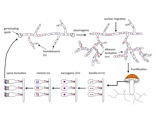
Everything you learned about sex- forget it, because mushrooms do it differently. For starters, after finding and fusing with an appropriate partner (different mating types), the mated hyphae form a mycelial type called a heterokaryon in which the two mated nuclei remain spatially distinct yet undergo mitosis in tandem (Figure 1). This cell type is similar to diploidy yet much more dynamic because nuclei maintain autonomy and may behave selfishly. A primary goal of our research in this area is to understand whether and when nuclei of different mating type in a heterokaryon can act as individuals, behaving selfishly, cooperatively, or displaying sexual selection. In previous research during my postdoctoral stint in Hanna Johannesson’s lab, we used a species with large numbers of nuclei per cell (Figure 2) to show that nuclei typically compete with each other (James et al. 2008). Our research in this area also asks whether three or more genomes can form cooperative partnerships and the role of meiotic and mitotic recombination in neutralizing or facilitating nuclear competition/cooperation.
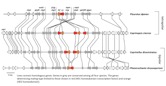
Mushrooms make a major departure from the classic battle of the sexes by the evolution of a multitude of sexes! Having more than two sexes or mating types allows a high level of promiscuity (or outbreeding efficiency), the causes and consequences of which are described in the paper (James 2015). Multiple mating types are generated by hyper-polymorphic mating-type genes that dictate inter-nuclear interactions. We are investigating what specific genetic changes to the mating-type genes lead to mating system evolution. We have developed molecular methods to analyze mating-type genes from non-model species and have explored the genetic changes responsible for the shift in mating system from one controlled by two loci (tetrapolar) to one controlled by a single mating-type locus (bipolar) in the fairies’ bonnets mushroom Coprinellus disseminatus and the white-rot fungus Phanerochaete chrysosporium. Results from both of these species suggest a convergent evolutionary mechanism specifically involving mutations to genes encoding pheromone receptors. In bipolar species, mating type is entirely controlled by a set of transcription factor proteins in a highly conserved region (Figure 3). We are now investigating the evolution of the signaling molecules that control nuclear communication and cooperation in bipolar mushroom species. We are also exploring how mating systems control inbreeding in mushroom populations, using species such as the bird’s nest fungi as a model system to relate spore dispersal mechanism, mating system, and population genetic structure.
Population genetics of chytridiomycosis
Chytridiomycosis is a disease of amphibians implicated in amphibian population declines in several regions throughout the world. Its agent, Batrachochytrium dendrobatidis (Bd), is a fungus that invades the epidermis of the animal and death is presumably caused by asphyxiation, osmotic imbalance, or toxin production. Chytridiomycosis is an emerging infectious disease because it is newly described and has created a documented wave of decline among sensitive populations. We are trying to answer two unresolved questions regarding the population genetics of the species: 1) what are the genetic origins of the disease and how has the disease been spread? 2) why has the disease spread so quickly despite some species being highly resistant? Our recent focus has been in the Atlantic Forest of Brazil where we have uncovered a genetically divergent and putatively endemic genotype of Bd (Schloegel et al. 2012) which seems to be less virulent and competitive than the global genotype (Jenkinson et al. 2018; Greenspan et al. 2018). This work is being done with collaborators at Cornell U. (Kelly Zamudio), U. Maine (Joyce Longcore), U. Alabama (Gui Becker) and UNICAMP in Brazil (L. Felipe Toledo, Domingos Leite). Our goals are to understand the distribution of endemic and pandemic Bd genotypes in the AF, how they relate to the distribution of the invasive North American bullfrog, and what threat the disease poses to endangered species.
Phylogeny of ancient fungal lineages
The “Tree of Life” is a major organizing principle for all of biological research and teaching. A major gap in our understanding of the Tree of Life is the order of the earliest branching lineages in the Kingdom Fungi. These groups include relatively morphologically simple, protist-like forms, including the Chytridiomycetes, microsporidia, and the recently described group Cryptomycota/Rozellomycota. We are researching the phylogeny of these basal lineages using genome sequence data. Through an NSF sponsored research project involving many mycology groups throughout North America (AFTOL) we reconstructed the history of the earliest fungi as a grade of zoosporic (motile spore) species, implying an aquatic habitat for early fungi (Figure 4). The loss of spore motility appears to have happened several times, each loss coincident with an innovation in spore dispersal and a major radiation of terrestrial fungi. As with the systematic treatments of other groups, the phylogeny of basal fungal lineages is a continual work in progress. We are currently using single cell genomics and metagenomics to understand the phylogeny and ecological function of members of Cryptomycota through an NSF sponsored project in collaboration with Igor Grigoriev at the Joint Genome Institute (DOE).