Development of Systematic Methodology to Deduce Interfacial Structures of Peptides/Proteins
The behavior of proteins and peptides at interfaces plays important roles in many applications and research fields such as biomaterials, marine antifouling coatings, biosensors using surface immobilized enzymes, membrane proteins, antibody drugs, antimicrobial peptides etc.
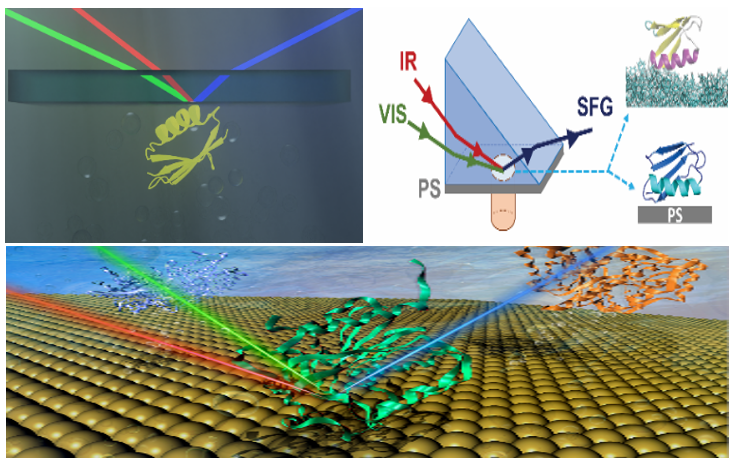
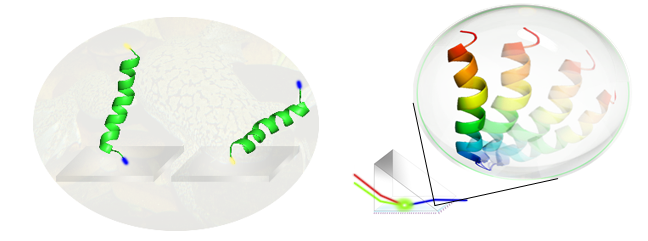
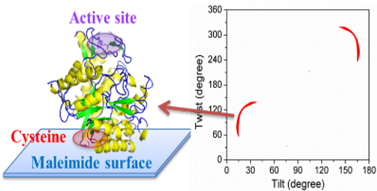
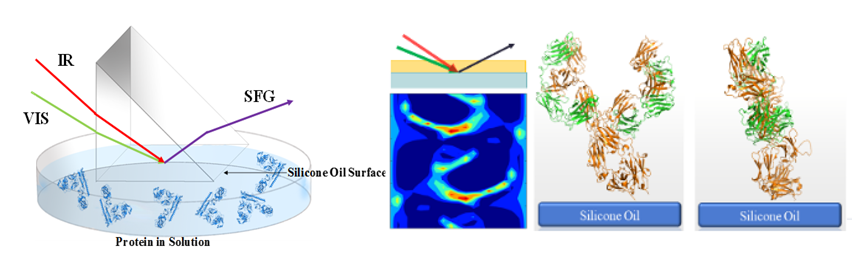
We are developing systematic methodology to deduce interfacial conformation and orientation of peptides and proteins.
This methodology is a combined approach with SFG measurements, Hamiltonian spectra calculation, spectral matching between experimental and calculated spectra, molecular dynamics simulations, and isotope labeling.
This method is widely applicable to study peptides and proteins at interfaces such as membrane proteins, surface immobilized enzymes and peptides, membrane associated antimicrobial peptides, antibody drugs on surfaces, physically adsorbed proteins, and adhesive proteins/peptides adhered to surfaces, etc.